Vascular Vectorization
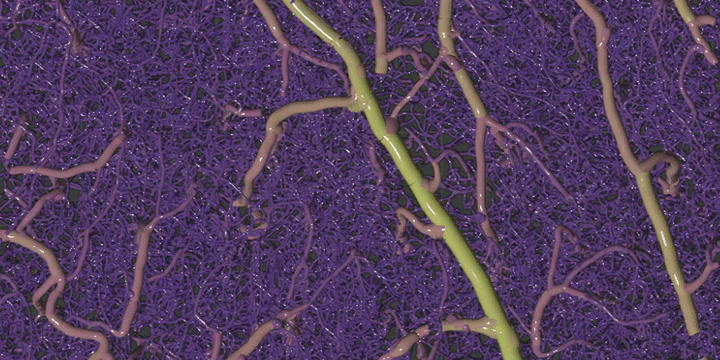 3D rendering of vectorized geometry
3D rendering of vectorized geometryRemarkably little is known about the plasticity of microvasculature in the adult brain because of the barriers to acquisition and processing of in vivo images. However, this basic concept is central to the field of neuroscience, and its investigation would provide insights to neurovascular conditions such as Alzheimer’s, diabetes, and stroke. We are developing the pipeline to image, map, and monitor the capillary blood vessels over several weeks to months in healthy mouse brains. One of the major challenges in this workflow is the process of extracting complex capillary networks from raw volumetric microscope images. This challenge is exacerbated by in vivo imaging constraints such as low resolutions, poor image quality, and contrast agents that highlight different features (e.g. plasma or endothelial labels). To confront these issues, we developed a general-purpose software method, Segmentation-Less, Automated, Vascular Vectorization (SLAVV), to extract vascular network maps from low quality images, enabling researchers to better quantify the vascular anatomy portrayed in their datasets.
).](/project/vascular-vectorization/statistics_huaac1c2c91e5cdb336b4347aa93399e96_858407_d4b2e726c8db2f3ac0cbbbfd780f5613.webp)